Seamless Cloning Solutions – Precision, Efficiency, and Flawless Results
"Achieve Perfect DNA Cloning with Our Excellent Seamless Cloning Kits"
----Fast, error-free cloning with ultra-high efficiency for your research needs
1. Introduction
Seamless cloning has become an indispensable tool in contemporary molecular biology, enabling precise DNA assembly without unwanted scars or restrictions. Our advanced cloning kits eliminate the limitations of traditional methods, offering unmatched accuracy, speed, and versatility for your cloning workflows.
Our Seamless Cloning Technology eliminates these frustrations by delivering:
✔ Flawless DNA assembly – No scars, no unwanted base additions.
✔ >95% cloning efficiency – Far higher than traditional ligation (often <30%).
✔ No restriction enzymes needed – Skip tedious digestion and purification steps.
✔ One-step, 5~15-minute reaction – From PCR product to ready-to-transform DNA in minutes, not days.
Whether you’re building complex vectors, assembling synthetic genes, or performing high-throughput cloning, our solution ensures perfect results the first time—accelerating your research without compromise.
Key Improvements Over Traditional Methods

2. How It Works
Design – Prepare your insert and vector with overlapping sequences (no specific sequences required).
Mix – Combine DNA fragments and vector with our master mix in one tube.
Transform – High-efficiency competent cells ensure reliable results.
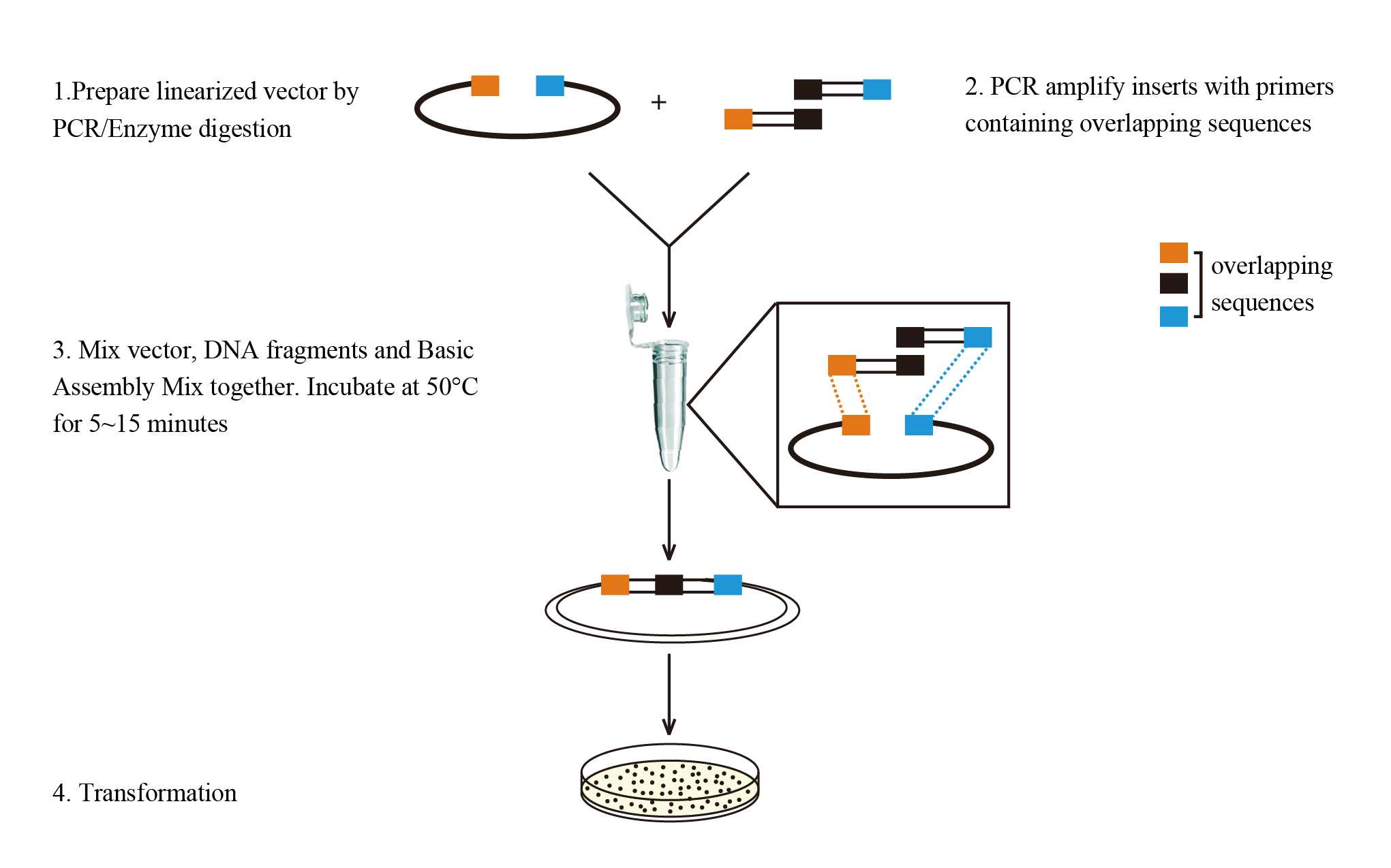
3. Our products
pEASY®-Uni Seamless Cloning and Assembly Kit (CU101)
▷ Fast and high efficiency
▷ Seamless assembly of 1-7 fragments
pEASY®-Basic Seamless Cloning and Assembly Kit (CU201)
▷ Fast and high efficiency
▷ Seamless assembly of 1-5 fragments
4. Why Choose Us?
Validated Performance – Trusted by leading research institutes and biotech firms.
Comprehensive Support – Detailed protocols, Seamless Cloning Primer Design Tool, Troubleshooting Guides, and expert assistance.
Single-Fragment Insertion with pEASY®-Uni Seamless Cloning and Assembly Kit (CU101)

PCR fragments of different sizes (50 bp, 2 kb, 5 kb, 8 kb) are inserted into the PG vector respectively, the ligation results are analyzed by endonuclease digestion. The results show that PCR fragments of different sizes are correctly ligated into the PG vector.
Multi-Fragment Insertion with pEASY®-Uni Seamless Cloning and Assembly Kit (CU101)

PCR fragments of different sizes (300 bp, 600 bp, 900 bp, 1200 bp, 1500 bp, 1800 bp, with NdeI restriction sites designed at both ends of the fragment) are mixed and inserted into
the pUC19 vector, the ligation results are analyzed by NdeI restriction enzyme digestion.The results show that fragments of different sizes are correctly ligated into the pUC19 vector.
Single-Fragment Insertion with pEASY®-Basic Seamless Cloning and Assembly Kit (CU201)

PCR fragments of different sizes (50 bp, 2 kb, 5 kb, 8 kb) are inserted into the PG vector respectively, the ligation results are analyzed by enzyme digestion. The results show that PCR fragments of different sizes are correctly ligated into the PG vector.
Multi-Fragment Insertion with pEASY®-Basic Seamless Cloning and Assembly Kit (CU201)

PCR fragments of different sizes (600 bp, 900 bp, 1200 bp, 1500 bp, with NdeI restriction sites designed at both ends of the fragment) are mixed and inserted into the pUC19 vector, the ligation results are analyzed by NdeI restriction enzyme digestion. The results show that fragments of different sizes are correctly ligated into the pUC19 vector.
5.Specifications
Product Name | Cat.No | Specifications |
CU101-01 | 10 rxns | |
CU101-02 | 20 rxns | |
CU101-03 | 20 rxns | |
CU201-02 | 20 rxns | |
CU201-03 | 60 rxns |
6.Related Products
Products | Product Name | Cat.No |
Nucleic Acid Purification | EE101 | |
| EE112 | ||
| ER302 | ||
| ER111 | ||
| ER501 | ||
| EP101 | ||
EasyPure® Plasmid MiniPrep Kit | EM101 | |
EasyPure® Quick Gel Extraction Kit | EG101 | |
RT-PCR | TransScript® One-Step gDNA Removal and cDNA Synthesis SuperMix | AT311 |
PCR SuperMixes | AS111 | |
| AS231 | ||
Modifying Enzymes | GD111 | |
Cloning Compent Cells | CD501 | |
DNA Molecular Weight Standards Related Products | GS101 |
Citations
pEASY® -Uni Seamless Cloning and Assembly Kit (CU101)
• Shi J Y, Mei C,Ge F Y, et,al. Resistance to Striga parasitism through reduction of strigolactone exudation[J] .Cell, 2025, (IF 45.5)
• Xu J, Liang Y, Li N, et al. Clathrin-associated carriers enable recycling through a kiss-and-run mechanism[J]. Nature Cell Biology, 2024. (IF 17.3)
• Wang J, An Z, Wu Z, et al. Spatial organization of PI3K-PI (3, 4, 5) P3-AKT signaling by focal adhesions[J]. Molecular Cell, 2024. (IF 14.5)
• Wang H, Yang J, Cai Y, et al. Macrophages suppress cardiac reprogramming of fibroblasts in vivo via IFN-mediated intercellular self-stimulating circuit[J]. Protein & Cell, 2024. (IF 13.6)
• Bai X, Sun P, Wang X, et al. Structure and dynamics of the EGFR/HER2 heterodimer[J]. Cell Discovery, 2023.(IF 13.0)
• Du J, Huang Z, Li Y, et al. Copper exerts cytotoxicity through inhibition of iron-sulfur cluster biogenesis on ISCA1/ISCA2/ISCU assembly proteins[J]. Free Radical Biology and Medicine, 2023.(IF 7.1)
• Zhu L, Li B, Li R, et al. METTL3 suppresses pancreatic ductal adenocarcinoma progression through activating endogenous dsRNA-induced anti-tumor immunity[J]. Cellular Oncology, 2023.(IF 4.9)
• Xu Y, Zhu T F. Mirror-image T7 transcription of chirally inverted ribosomal and functional RNAs[J]. Science, 2022.(IF 44.7)
• Shi C, Yang X, Hou Y, et al. USP15 promotes cGAS activation through deubiquitylation and liquid condensation[J]. Nucleic Acids Research, 2022.(IF 16.6)
• Jin Q, Yang X, Gou S, et al. Double knock-in pig models with elements of binary Tet-On and phiC31 integrase systems for controllable and switchable gene expression[J]. Science China Life Sciences, 2022.(IF 8.0)
• Duan Y, Liu S, Gao Y, et al. Macrolides mediate transcriptional activation of the msr (E)-mph (E) operon through histone-like nucleoid-structuring protein (HNS) and cAMP receptor protein (CRP)[J]. Journal of Antimicrobial Chemotherapy, 2022.(IF 3.9)
• Li B, Zhu L, Lu C, et al. circNDUFB2 inhibits non-small cell lung cancer progression via destabilizing IGF2BPs and activating anti-tumor immunity[J]. Nature communications, 2021.(IF 14.7)
• Song N, Chen L, Ren X, et al. N-Glycans and sulfated glycosaminoglycans contribute to the action of diverse Tc toxins on mammalian cells[J]. PLoS Pathogens, 2021.(IF 5.5)
• Liu S, Zhang M, Bao Y, et al. Characterization of a highly selective 2 ″-O-galactosyltransferase from Trollius chinensis and structure-guided engineering for improving UDP-glucose selectivity[J]. Organic Letters, 2021.(IF 4.9)
• Liu S, Fan L, Liu Z, et al. A Pd1–Ps–P1 feedback loop controls pubescence density in soybean[J]. Molecular plant, 2020. (IF 17.1)
pEASY® -Basic Seamless Cloning and Assembly Kit (CU201)
• Zhang R, He Z, Shi Y, et al. Amplification editing enables efficient and precise duplication of DNA from short sequence to megabase and chromosomal scale[J]. Cell, 2024.(IF 45.5)
• Lei X, Huang A, Chen D, et al. Rapid generation of long, chemically modified pegRNAs for prime editing[J]. Nature Biotechnology, 2024.(IF 33.1)
• Zhang Y, Dai F, Chen N, et al. Structural insights into VAChT neurotransmitter recognition and inhibition[J]. Cell Research, 2024.(IF 28.1)
• Jiang C, Yu C, Sun S, et al. A new anti-CRISPR gene promotes the spread of drug-resistance plasmids in Klebsiella pneumoniae[J]. Nucleic Acids Research, 2024.(IF 16.6)
• Ye D, Shao Y, Li W R, et al. Characterization and Engineering of Two Highly Paralogous Sesquiterpene Synthases Reveal a Regioselective Reprotonation Switch[J]. Angewandte Chemie International Edition, 2024.(IF 16.1)
• Chen D, Song Z, Han J, et al. Targeted Discovery of Glycosylated Natural Products by Tailoring Enzyme-Guided Genome Mining and MS-Based Metabolome Analysis[J]. Journal of the American Chemical Society, 2024.(IF 14.4)
• Zuo F, Jiang L, Su N, et al. Imaging the dynamics of messenger RNA with a bright and stable green fluorescent RNA[J]. Nature Chemical Biology, 2024.(IF 12.9)
• You L, Omollo E O, Yu C, et al. Structural basis for intrinsic transcription termination[J]. Nature, 2023.(IF 50.5)
• Huang J, Yang L, Yang L, et al. Stigma receptors control intraspecies and interspecies barriers in Brassicaceae[J]. Nature, 2023.(IF 50.5)
• Bai X, Sun P, Wang X, et al. Structure and dynamics of the EGFR/HER2 heterodimer[J]. Cell Discovery, 2023.(IF 13.0)
• Zou Z P, Du Y, Fang T T, et al. Biomarker-responsive engineered probiotic diagnoses, records, and ameliorates inflammatory bowel disease in mice[J]. Cell Host & Microbe, 2022.(IF 20.6)
• Wan W, Dong H, Lai D H, et al. The Toxoplasma micropore mediates endocytosis for selective nutrient salvage from host cell compartments[J]. Nature communications, 2023.(IF 14.7)
• Jiang L, Xie X, Su N, et al. Large Stokes shift fluorescent RNAs for dual-emission fluorescence and bioluminescence imaging in live cells[J]. Nature Methods, 2023.(IF 36.1)
• Liu R, Yao J, Zhou S, et al. Spatiotemporal control of RNA metabolism and CRISPR–Cas functions using engineered photoswitchable RNA-binding proteins[J]. Nature Protocols, 2023.(IF 13.1)
• Liu R, Yang J, Yao J, et al. Optogenetic control of RNA function and metabolism using engineered light-switchable RNA-binding proteins[J]. Nature biotechnology, 2022.(IF 33.1)



